Current Research
Funded Projects
Stability of Regulators in Embryonic Development (Epistemodevo)
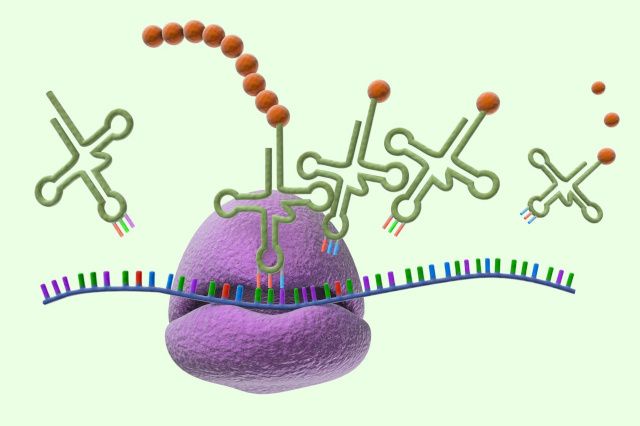
Developmental biology has largely aimed at understanding how genes and gene regulatory networks (GRNs) shape tissues, organs and organisms. But what happens if we add an additional layer of regulation after gene transcription and translation (or “hypogenetic” regulation) to these models? This project, a collaboration with three developmental biologists (René Rezsohazy, Françoise Gofflot, and Frédéric Lemaigre), includes an experimental aspect, characterizing a neglected layer of molecular regulations taking place at the level of proteins (transcription factor stability), and a philosophical aspect, evaluating the importance of and changes in practices of model building and idealization in describing complex developmental processes. In particular, we will explore the extent to which adding this extra layer of protein regulation changes the theoretical and experimental presuppositions of the proposed biological work, investigating whether the whole is, potentially, more than the sum of its parts.
You can find an archive of our former financed projects here.
Research Areas
Foundations of natural selection

The primary research project of our lab has to do with what we call “conceptual foundations” work in evolutionary theory. How should we – in general, as opposed to in specific natural populations – think about the causal structure of evolution by natural selection? What role do various concepts like fitness, selection, drift, and mutation play? In particular, evolutionary theory is filled with invocations of probability and statistics. But how should we understand this? How many sources of chance are there, and how should they be interpreted?
To explore this question, work in our group has offered new analyses of the concept of fitness, on other key concepts like genetic drift, and is currently exploring the nature of biodiversity and the basic causal structure of evolutionary theory.
History of biology

If we are to understand the current role of chance in evolutionary theory, we must also understand the history that led to chance having played that role. Our group thus has an extensive historical focus, considering the ways in which Darwin understood chance, and, moving slightly farther forward in history, how the biologists who first introduced statistical reasoning into evolutionary theory – particularly Francis Galton, Karl Pearson, and W.F.R. Weldon – conceptualized their work. Seeing why these biologists believed (several decades before the development of “modern” evolutionary theory) that chance and statistics were integral to their work remains an enormously rewarding effort.
Work in our lab focuses on the collaboration between W.F.R. Weldon and Karl Pearson, particularly their approach to natural selection and inheritance; work leading to, among other things, the publication of a book, The Rise of Chance in Evolutionary Theory. We work both with publicly available and archival materials, and welcome students and collaborators whose primary interest is in the history of science.
Digital humanities
One important task for philosophers of biology is to be able to “take the pulse” of the biological literature. With regard to our research questions, for example, what are biologists currently saying about historical contingency? Or about fitness? Answering such questions is enormously difficult, in no small part thanks to the magnitude of the biological literature. Thousands of articles are published every day – we clearly cannot expect to answer broad-scale, general questions about biology journals without the aid of computer analysis.
To that end, our group has worked significantly on textual analysis of scientific journal articles. Formerly, this took the shape of the Sciveyor project, though now we develop our own small solutions in-house. We are excited to work with anyone interested in the digital humanities on increasing their capabilities.
